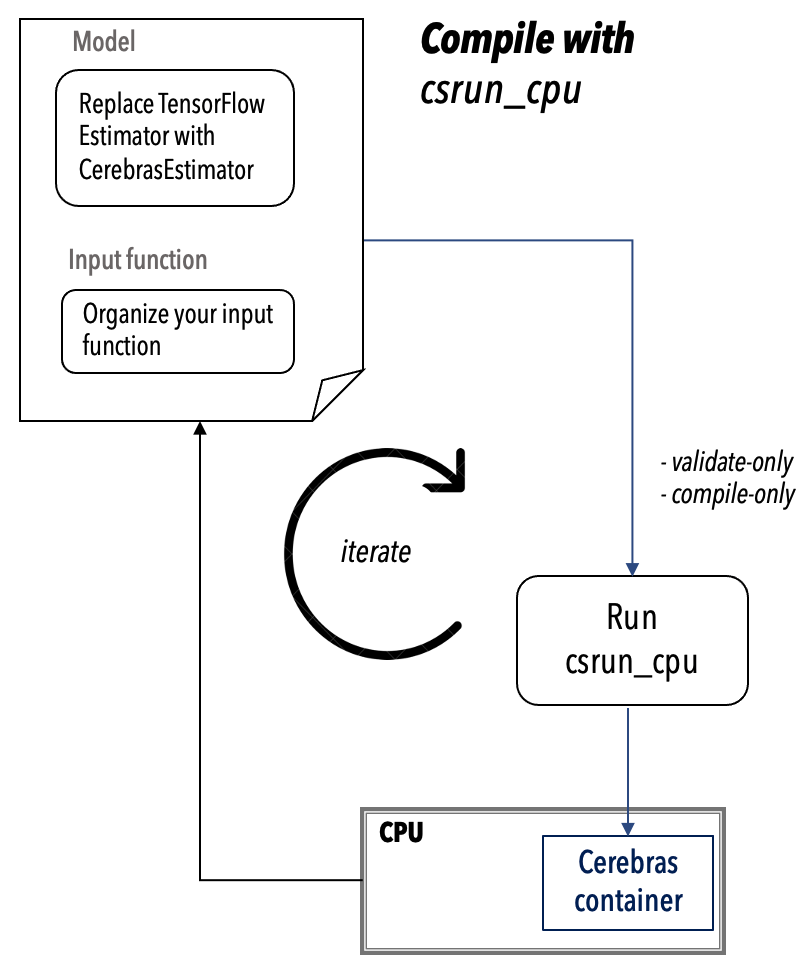
The csrun_cpu Script#
Use the csrun_cpu script to compile your model on a CPU node, before running the model on the CS system.
Note
Slurm wrapper scripts (csrun_wse and csrun_cpu) may be customized for your particular environment by your sysadmins and may look different than what is shown below. Check whether your Sysadmin’s local documentation is available and whether there are any special instructions for your CS-2.
Configuring csrun_cpu#
If you are following pipelinedmodels using the Slurm/Singularity workflow#
Before you can use csrun_cpu, the system administrator must configure this script with proper variables. Follow the below guidelines:
The scripts
csrun_cpuand thecsrun_wseare used together in the Cerebras ML workflow. Hence, place these scripts in a commonly accessible location. These are Bash executable scripts, so you can place them where other executables are located.Ensure that all the scripts are executable (use
chmod +x csrun_cpu, for example).Ensure that the location of the scripts is included in the PATH variable.
To confirm that you have the correct set up, run
csrun_cpu pythonon a command line. This should launch a Python interpreter inside the Cerebras Singularity Container.Edit the
csrun_cpuscript and set the variables in it. See the following code section in thecsrun_cpuscript where these variables are located:# All that needs to be set by system admins for different systems is here ######################################################################## # sif image location SINGULARITY_IMAGE= # Comma seperated string of directories to mount. # ex: MOUNT_DIRS="/data/,/home/" # Note that the current directory is always mounted. So no need to add ${pwd} MOUNT_DIRS= # Default slurm cluster settings (must be set) DEF_NODES= DEF_TASKS_PER_NODE= DEF_CPUS_PER_TASK= #### More slurm configurations (recommended but not required) ##### # The name of the GRES resource. GRES_RESOURCE= # The GRES node associated with the gres resource GRES_NODE= ########################################################################
The values of these variables depend on your location of the SIF image, default Slurm configurations, the default directories to mount, and so on. These variable settings will be used by the Cerebras compiler when running jobs on the CS system. Consult Cerebras support if you need help setting the Slurm defaults.
Important
Specifying GRES_RESOURCE and GRES_NODE avoids conflicts when scheduling CS-2 jobs using slurm. Consult with the system administrator and Cerebras support for the configuration of these environment variables.
If you are running the pipelinedmodels in the Kubernetes workflow#
The users scripts rely on a set of default options that are provided by system admins. These defaults must be placed in a .yaml file and the filepath must be passed as --admin-defaults argument to the scripts.
Here’s an example of what a .yaml file with these defaults may look like:
# Default K8s cluster settings (must be set) DEF_NODES: 2 DEF_TASKS_PER_NODE: 5 DEF_CPUS_PER_TASK: 16 # cbcore image location CBCORE_IMAGE: "/default/path/to/cbcore" # Comma seperated string of directories to mount. e.g., "/data/,/home/" # Note that the current directory is always mounted. So no need to add PWD MOUNT_DIRS: "/default/some/mount/dir,/another/mount/dir" # Certificate path to access the cerebras cluster CERTIFICATE_PATH: "/default/path/to/certificate" # Cerebras Cluster Server to connect to CLUSTER_SERVER: "cluster-server.cerebras.com"
csrun_cpu#
>csrun_cpu --help
Usage: csrun_cpu [--help] [--alloc_node] [--mount_dirs] [--use-sbatch] command_to_execute
...
...
...
Description#
Runs the given <command_to_execute> inside the Cerebras environment on a CPU node.
Arguments#
command_to_execute: A user command, such aspython run.pyorbash, that is executed inside the Cerebras container on a CPU node.--alloc_node: (Optional) Set this toFalseif you do not wish to reserve the CPU node exclusively to execute <command_to_execute>. Default isTrue. This applies only for Slurm and not for the Kubernetes workflow.--mount_dirs: (Optional) String of comma-seperated paths to mount in addition to the standard paths listed incsrun_cpu. Default is an empty string, i.e., only paths listed incsrun_cpuare mounted.--use-sbatch: (Optional) Adding this flag will submit a batch script to slurm to execute <command_to_execute>. sbatch will immediately exit after submitting the script. The script will stay on the slurm queue of pending jobs until resources are allocated. This applies only for Slurm and not for the Kubernetes workflow.
Important
You must compile on a CPU node from within the Cerebras Singularity container. The csrun_cpu script ensures that your compile is run within the Cerebras Singularity container.
Examples#
csrun_cpu --mount_dirs="/data/ml,/lab/ml" python run.py --mode=train --validate_only
Mounts /data/ml and /lab/ml in addition to the default mount directories and then executes the command
python run.py --mode=train --validate_only, which runs validation inside the Cerebras container on a CPU node.
csrun_cpu --alloc_node=True --use-sbatch python run.py --mode=train --compile_only
Submits a sbatch job to slurm that reserves the whole CPU node and executes the command
python run.py --mode=train --compile_only, which runs the compilation inside the Cerebras container on the reserved CPU node.
csrun_cpu python
Launches a Python interpreter inside the Cerebras container on a CPU node.
Validate only#
With validate_only mode, the CGC will run in a lightweight verification mode. In this mode, the CGC will only run through the first few stages of the compilation stack, up through kernel matching.
This step is very fast and will allow you to quickly iterate on your model code. It enables you to determine if you are using any functionality that is unsupported by either XLA or the Cerebras stack.
Here is an example command:
csrun_cpu --mount_dirs="/data/ml,/lab/ml" python run.py --mode=train --validate_only
The above command mounts
/data/mland/lab/mldirectories, in addition to the default mount directories, and then executes the Python command:python run.py --mode=train --validate_only. The Python command validates whether your training graph is supported by the Cerebras software. Thiscsrun_cpucommand will automatically spin up the Cerebras container on a CPU node to run this step.
A successful run in this mode validates the following:
Your model code is fully CS-compatible.
Your model correctly translates through XLA, and
Your model is supported by the available Cerebras kernels.
Note
A successful run in the validate_only mode does not mean that your model is guaranteed to compile. Compilation may still fail in lower-level stages of the Cerebras stack. However, any errors you reach beyond this stage are issues not with your model, but with the Cerebras software stack and should be reported to the Cerebras Support Team.
Compile only#
With compile_only mode, the CGC will perform full compilation through all stages of the Cerebras Software Stack and generates a CS system executable. Note that it will not run the executable on the CS system in this mode. However, when the compile_only mode is successful, your model is likely to run on the CS system.
Here is an example command:
csrun_cpu --alloc_node=True python run.py --mode=train --compile_only
The above command reserves the whole CPU node and executes the Python command:
python run.py --mode=train --compile_only. The Python command compiles a mapping of the training graph on the reserved CPU node.
You can save time in your workflow by using the compiled artifacts from the compile_only session in your subsequent execution of this network on the CS system. This allows you to skip the compile step on the CS system, thereby saving you time in your workflow.
Note
Note that you must use csrun_wse script to run on the CS system.
Hardware resource recommendations#
When using the compile_only option to compile models within the Cerebras environment, we recommend 64GB of memory and at least 8 cores as a minimum requirement. Make sure that these resources are dedicated to the compile and are not shared.
For example, when you run from within a Cerebras Singularity container, you compile a model with the compile_only option, as shown below:
csrun_cpu python run.py --mode=train --compile_only
then, if the hardware resources are less than the above minimum, the compile may fail with the following error:
cerebras.cigar.stack.CerebrasStackError: [Cerebras Internal Error (source "plangen")]
Compilation internal error at stage plangen.
[...]
terminate called after throwing an instance of 'std::bad_alloc'
what(): std::bad_alloc
Validate and compile outside the CS system cluster#
To validate and compile from outside the Cerebras cluster, do not use Slurm to invoke the standard Singularity container. Instead, directly launch the Singularity container interactive shell with the proper path to the Cerebras Singularity image:
singularity shell --cleanenv -B {data folders to attach} {path/to/singularity}/cbcore-[version-number].sif
# Full compile
csrun_cpu python run.py --mode=train --compile_only
# Validation only
csrun_cpu python run.py --mode=train --validate_only
Important
We recommend that you initially iterate on your model with validate_only option for the run.py.
Sbatch mode#
The default behavior of csrun_cpu uses srun. With srun, slurm will allocate resources and csrun_cpu will exit once the slurm job is finished. By using the flag --use-sbatch, csrun_cpu submits to slurm a batch script to execute the command <command_to_execute> using sbatch. sbatch will immediately exit after submitting the script. The script will stay on the slurm queue of pending jobs until resources are allocated.
The command use will be stored as the file CS_<date>.log and the standard output and standard error will be stored as CS_<date>_<slurm_job_id>.out.
If a CS-2 dedicaded CPU node is specified using GRES_NODE, then csrun_cpu will avoid using this node for compilation tasks.